EDITORS NOTE: (November 16, 2017) The New York Times writes that a consensus is developing among scientists that deploying gene drives is too risky for field trials.
There is no genetic code. Not really. Not in the way most people think. Seasoned, sensible geneticists know it’s true.
Unfortunately, a few immature biologists don’t believe it. They are developing “gene drive” technologies that they hope will enable them to reliably and permanently alter a fragment of “the code” in any life-form that reproduces sexually — to guarantee that the altered piece of “code” will be transmitted to the next generation 100% of the time into perpetuity.
NOTE TO READERS: November 22, 2019: This essay is the longest on the website. To help readers navigate, The Editors asked Billy Lee to add links to important subtopics. Don’t forget to click the up arrow on the right side of the page to return to top.
1 — Gene Drivers
2 — Genetic Code. What is it?
3 — Bases
4 — Animal DNA
5 — Mitochondria & Bacteria
6 — DNA analogy
7 — Genes?
8 — RNA
9 — Ribosomes
10 — Nucleic acid
11 — Scale of DNA
12 — Rosalind Franklin
13 — Genomic weather
14 — Jurassic Park
15 — Clones
16 — Enzymes
17 — XNA
18 — CRISPR
19 — Non-DNA life
20 — Dark DNA
21 — Junk DNA
22 — Why species?
23 — Acknowledgment
Deployment of gene-drive technology means that an altered fragment of genetic code can be “injected” into a species, for good or ill, which is permanent and will over a few generations become universal — unable to be suppressed or removed regardless of any natural selection pressures whatsoever — until the end of time.
The changes caused by gene drivers takeover every individual in any species that has been targeted for modification. It takes about 10 generations, give or take. With insects, we’re talking a couple of years; plants, a decade maybe; humans, 300 years or so.
Gene drivers are all about changing an entire species forever and permanently — not just one individual with a genetic disorder, for example, or one generation of plants for another. It’s a higher level of intervention than conventional gene therapies and modifications.
A screw-up can extinguish a species in a relatively short period of time is how I see the danger. Worse, according to the scientists cited by the NYTimes, these genes will migrate into ecological niches, where they will force unintended consequences to the biosphere; worse still, given sufficient time “good” genes are likely to jump species, where they will wreak havoc.
EDITORS NOTE: On 18 September 2020 the website science-news service phys.org published an article titled Biologists Create New Genetic Systems to Neutralize Gene Drives.
According to the article:
The first neutralizing system, called e-CHACR (erasing Constructs Hitchhiking on the Autocatalytic Chain Reaction) is designed to halt the spread of a gene drive by “shooting it with its own gun.” e-CHACRs use the CRISPR enzyme Cas9 carried on a gene drive to copy itself, while simultaneously mutating and inactivating the Cas9 gene.
The system can in principle be placed anywhere in the genome.
The second neutralizing system, called ERACR (Element Reversing the Autocatalytic Chain Reaction), is designed to eliminate the gene drive altogether. ERACRs are designed to be inserted at the site of the gene drive, where they use the Cas9 from the gene drive to attack either side of the Cas9, cutting it out. Once the gene drive is deleted, the ERACR copies itself and replaces the gene-drive.
Both systems have been tested in the lab at a molecular level. The developers have not yet demonstrated that a gene drive screw-up gone wild can be pulled back and eliminated by these systems.
The good news is that scientists are working on the problem. The bad news is that the existence of infant, untested technologies might tempt some to release a gene drive into the wild that will ultimately prove intractable.
EDITORS NOTE: On 6 March 2021 the website science-news service phys.org published an article titled New ‘split-drive’ system puts scientists in the (gene) driver seat. The piece describes new split-gene-drive technology that promises to degrade over several generations to permit engineered genomes to evolve under the rules of natural selection. Such a system, if safely deployed, would help to prevent collapse-of-species and other bad consequences when mistakes are made.
If anybody doesn’t understand what they just read, they shouldn’t worry. By the end of this essay, they will fully grasp why the scientists pursuing this course might be dangerously eager to unleash genetic pollutants that may kill us all — because their love of science makes it difficult to restrain themselves.
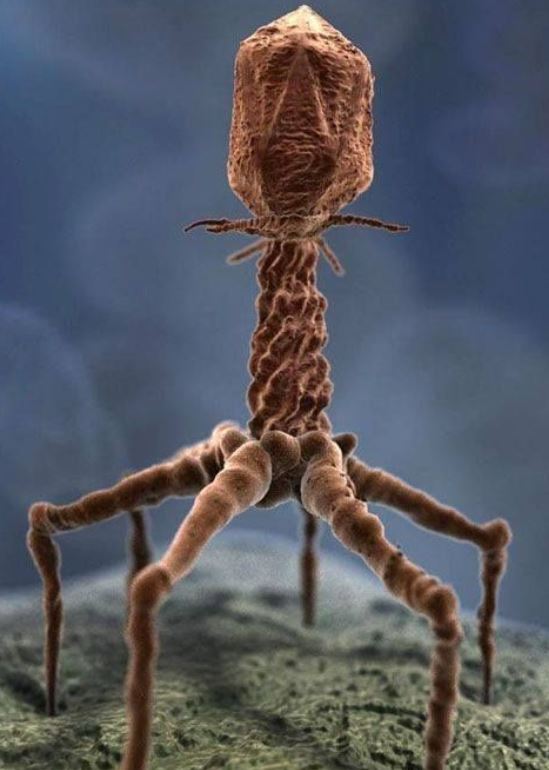
These smart people (some might be prodigies for all I know) plan to use gene drivers to exterminate vermin and eradicate insect-borne disease, for starters. They plan to make it impossible for agricultural pests to develop resistance to pesticides.
It all sounds great. But so did using the by-products of nuclear bombs as an energy source for our cities. Ask Japan how their state-of-the-art nuclear energy program turned out. Ask about Fukushima.
Read Nuclear Power and Me and 47 TONS to learn more.
Gene-drive technologies are an existential threat to the long-term survival of life on Earth — like those tens-of-thousands of plutonium-loaded thermonuclear missiles, which a number of countries have buried a few hundred feet below the surface of the earth.
The warheads on these missiles are going to rot someday, because no one can take care of them forever, and we can’t get rid of them. Their poisons — the most lethal known; a speck of plutonium dust can kill any human who ingests it — will leach into the soils; over thousands of years percolating plutonium will kill everything.
Genes — bad ones (or very good ones that turn out bad; oops!) — genes that can never die; genes that can’t be suppressed by natural selection; genes that are always passed on to the next generation under every conceivable scenario and every possible pairing of mates (no matter how mismatched) present potential nightmare scenarios for any species that possess them. Errant gene drivers can extinguish some species in a matter of a few years.
It is distressing to think that smart, young adults — I can imagine some younger than 35 who possibly lack basic common-sense — does it matter how smart they are? — might right now be playing around with molecules of DNA they can’t possibly understand fully, because the molecules have a quantum side to their nature that can make their behavior unpredictable; even unknowable.
Young adults are messing around with very complicated structures and processes inside both molecules and cells that they can’t see, even with the best microscopes and the most sophisticated instruments. It’s possible that they might — even with the best intentions; the best lab protocols — screw things up big-time and possibly forever. We need maximum oversight over these researchers and the labs who employ them, now — not tomorrow or next year.
Click on this link to an essay in the science journal Nature, which addresses the issue of gene-driver risk and its management. It is written by someone who seems, at least to my mind, to suggest remedies that are insufficiently robust.
Every biologist knows that “genes” have a mysterious way of migrating between species, crossing boundaries, and behaving unpredictably. They have a way of escaping confinement structures. If folks don’t understand why, what are they doing playing around and calling it research?
Editors Note (May 27, 2017): Here is a link to a May 17, 2017 article in Science News about the role of jumping genes in the expression of genomes, which may be of interest to some readers.
People have claimed for years that recombinant DNA pathogens, retro-viruses and yes, AIDS, escaped from rogue laboratories. Does anyone know for sure? If they do, they aren’t saying.
Anyway, I urge readers to relax for now in the knowledge that they are about to learn some amazing things. I did, writing this essay. And please remember: I am a pontificator, not a scientist.
Links are provided to verify anything written in the essays that people may question. My pledge to readers is, as always, to be as accurate as possible and to correct mistakes should I discover them or find myself corrected by others.
Yes, I can be smart and write good too. Well, I’m trying anyway. Getting it right is important. It’s my highest priority. But sometimes I screw up — usually on some arcane technical detail in an essay about science.
Sometimes the science changes and new facts emerge. When I first published this essay in May 2016 everyone thought the galaxies in the universe numbered 200 billion. As I add this note in May 2018, analysis of the latest pics from telescopes in space suggests that the number of galaxies is closer to two-trillion.
My pledge is to keep my essays up-to-date and to learn enough to fix screw-ups that might be caused by simple ignorance. Readers can help fix errors by sending corrections in comments. Errors in the text will be fixed immediately.
So far, we’ve been lucky. The number of errors identified is amazingly few. In one doozy, I published the picture of a well-known British actress (well-known in Britain) and said it was Rosalind Franklin, the X-ray crystallographer famous for making the images that hinted at the spiral staircase structure of DNA.
Celebrities in Europe (that is, in England) were kind enough to take the time to inform me how wrong I was. I appreciated the feedback and was grateful they didn’t kill me.
I’m joking. The Brits are the kindest and best-behaved people on Earth. I’ve spent enough time in London to know.
The genetic code that everyone talks about lives inside tiny spaces; one way to think about it is to imagine that it lives inside little rooms packed to the ceiling with sacks — bitty-bags stuffed full of strung-together bases — hiding in the center of every cell of every plant and animal (or disbursed throughout the cell in the case of most one-celled microbes).
It isn’t a code at all. It’s a reservoir; an inventory; a collection of templates — most broken into pieces; separated and scattered among the dozens of spiral tentacles in the vast aperiodic-crystal known as DNA.
DNA isn’t exactly a crystal either; not really. Crystals are structures made from regular (periodic) arrangements of molecules. DNA, on the other hand, is a molecule, and it is constructed from strings of chemical bases. It’s found in bundles alongside other DNA molecules. These bundles are called chromosomes.
During part of a cell’s life cycle these DNA strings can sometimes be found tightly wound around little pieces of protein like fishing lines around spinning-reels. It’s a configuration that makes them compact; less intrusive — but easier to see under a microscope when they have been stained.
Inside any particular cell each chromosome of DNA is in some ways a little like a snowflake; within a single cell, no two chromosomes are the same; no two are alike, not even close. But every cell in the body contains the same group of chromosomes; the chromosomes in each body-cell are identical to the chromosomes in every other body-cell, right? It’s not hard to understand.
Bases, by the way, (in case someone might be wondering) are chemical substances that turn into salts when acids are poured over them. Many bases exist in nature, but only four (nucleobases) are found in DNA.
These four bases are essentially one or two rings of nitrogen and carbon atoms with ammonia and vinegar-like side chains attached. Here are links for anyone who wants to look them up: adenine – thymine ; and cytosine – guanine.
A fifth base, uracil substitutes for thymine in RNA, which is a vast assortment of short, single-stranded, DNA-like segments — the cell’s worker-ants who enable cells to perform their many functions. RNA builds genes, moves stuff around, and dramatically speeds up cell chemistry by catalyzing thousands of processes.
Not to digress, but NASA found uracil on the surface of Saturn’s moon Titan in 2012 during a fly-by. It’s something to wonder about.
Caffeine is molecularly similar to the bases adenine and guanine. Bases taste bitter — like the caffeine in coffee. Acids, on the other hand, taste sour — like vinegar. Combine bases and acids (bitter and sour) chemically to make salts, which are substances that taste like the ocean.

Forgive me for starting simple. Life-sciences are the most complicated sciences of all.
What adds to the difficulty is that in most animals (and all people) the DNA involved in sexual reproduction is configured differently than the DNA in body-cells. It exhibits behaviors a little less like those found in other cells.
In this essay we are talking about animal DNA, usually human, in body-cells — somatic cells; and we are talking about protein production.
A single DNA group inside certain human body-cells like liver cells and stem cells (while they undergo the process of replicating and dividing) is composed of ninety-two large molecules called DNA strands, which together warehouse the six billion base-pairs that will populate the genomes of two daughter cells.
During the short-lived interval when cells divide and replicate, dozens of molecules and billions of bases gather themselves into the configuration of chromosome-pairs peculiar to people, which some of us learned about in high school biology.
Most of the time (90% of it anyway) DNA doesn’t divide and multiply; it doesn’t organize itself into easy to recognize chromosomes; in all body-cells except stem cells and liver cells, DNA is the starting material for the making of proteins instead.
Making proteins is the only thing most body-cells do; it starts in the DNA molecules and is the subject of this essay. Cell division and replication is what stem cells do. Links will lead to those subjects for any who might want to learn more.
This essay is not about stem cells, which develop into any and every kind of somatic (body) cell and germ (reproductive) cell. As a pontificator who is not an expert on stem cells, my understanding is that — except for liver cells — somatic cells in mature adults don’t generally divide and reproduce themselves. That function is performed by stem cells, which start at conception and continue through life to replenish the human body.
Stem cells live inside the tissues of adults like seasonings inside cooked beef, is how I imagine it. Check me on this one, experts. Correct me in the comments section. All others read this link on cell differentiation first.
The main point is that human body-cells house 46 chromosomes (called chromatids when they are organized into 23 connected pairs), which contain six billion base-pairs. Reproductive (germination or germ) cells contain 23 unpaired chromosomes that store three billion base-pairs.
Confusing terminology constructed from the Greek language can create stumbling blocks for non-scientists, so I’m reluctant to go there. Terms like diploid, haploid, gamete, and zygote folks can look up and explore on their own. There’s enough that’s fascinating in English. Only tiny, digestible Greek lessons — sparsely sprinkled — will appear in this essay.
Besides unfamiliar vocabulary, another hard concept to grasp is this: inside every animal cell (and plant cell) are hundreds of DNA packed bundles (called mitochondria), where the DNA is not like the DNA in sex-cells or in body-cells either. The DNA in mitochondria matches what one might expect to find in another as yet undiscovered species of bacteria. It’s “coded” differently.
Yes, it’s weird, but there are explanations.
Most scientists today believe that a long time ago cells engulfed bacteria; these foreign migrants from another world (in terms of scale) were simply unable to escape.
Bacteria are small. A thimble-full of dirt can contain 50,000 species. It is amazing to learn that millions of species of bacteria exist in the soils and on the surfaces of plants on the earth.
Thirty-percent of cells in the human body are bacteria. They don’t weigh much, because they are small. It would require as many as 10,000 individuals of some strains to match in size just one of the microbes displayed in the illustration a few paragraphs above.
On average, though, a typical cell in an animal or plant can be visualized as having about 4,000 times the volume of a typical bacterium. The range of volume ratios varies widely, of course. Nothing is simple, especially in biology. Enough said.
Scientists named the trapped bacteria-like life-forms inside cells, mitochondria, after the Greek words for threaded granules; these granules make the cells they inhabit more robust, because they act like little batteries, boosting the energy in their adopted homes to help power the many tasks that cells do. Click the video link above for an animation by Harvard University that shows how it works.
Click the link above for an easy-to-understand animation of the overall structure of cells; or this link for a YouTube Video designed to transport viewers through an imaginary, animated world that makes real the complexity of a working, living cell.
And here is a link from Wikipedia, if anyone is confused about the numbers of bases and chromosomes in humans, as many folks seem to be, including myself, sometimes.
It’s confusing, because there are different “codes”, different cell-cycle phases, different collections of DNA molecules in body, germ, and stem cells — and I haven’t even mentioned enzyme catalysis or polymerases (and I’m not going to, either — not just yet anyway — because it will open a big can of worms I don’t want to deal with right now).
Don’t worry, we’ll get to some of it later after I’ve laid a little scaffolding.
But let me say this: without all this complexity, life forms as complicated as human beings would be impossible — codes or no codes.
The tools most people use to do science, especially physics, generally depend on mathematics and rigid, predictable rules. The life-sciences aren’t like that; not at all.
Should my essay devolve into complexity, readers are free to bail. I’m going to try to keep the mysteries of DNA understandable to non-technical people. Who knows if I’ll succeed or not?
DNA can be thought of as a collection of pouches or bags stuffed with billions of copies of four basic substances, called bases. DNA is like a roomful of holiday bags, each filled to the brim with four different kinds of unfinished toys like the ones in Santa’s workshops before Christmas.
Each of the four kinds of toys are strung together in-line, one after the other — in no discernible order — in long, tangled spirals. These spirals are unimaginably long, and there are many dozens of them.
The toys in the DNA sacks are unfinished, unpainted, and undecorated. They really don’t look much like toys at all. In this analogy, the four bases might be imagined as four simple blocks of wood, each a different shape and size. And like I said, there are billions of these blocks, at least in human cells.
Is DNA a big molecule? Yes, I already said that it was. It’s huge. But good luck to anyone who tries to find one. Good luck to anyone who tries to look at one. No one has ever seen any molecule. No matter how large, molecules are too small to see, even with microscopes; and that includes DNA molecules, the largest and most complex molecules in biology.
It takes a combination of high-energy light, amplification, and computer-generated algorithms to produce useful pictures of what scientists think molecules look like. A computer-generated image is not the same as a brain-generated image stimulated by the act of looking at reflected light with a pair of human eyes.
X-ray crystallography was the technique used first to unravel clues to the structure of DNA. From the data collected by crystallography, Linus Pauling shared in 1951 compelling ideas about what he thought the structure was, but he turned out to be wrong. He got protein structure right, but his description of DNA had subtle errors. A few years later, others came up with a structure that has thus far stood the test of time.
In soma (body) cells, forty-six molecules (strands or bags or sacks) of DNA contain the six billion base-pairs (or blocks) of the human genome. Most of the time these strands are loose and disorganized; a diffuse mass of hard-to-see chromatin. (Their form depends on what part of its period the cell cycle is in.)
It has to be this way for the worker elves of the cell to gain access to the bases (the unfinished toy blocks) upon which they will do their work. Only during the process of cell division do these forty-six molecules bind together and curl-up into the twenty-three chromosome-pairs that some readers may have learned about in high school biology class.

Researchers have technologies that can amplify what DNA molecules reveal, which they manipulate with computer algorithms to form fuzzy pictures that are helpful to highly trained analysts; but it’s the best they can do, visually.
An early theorist, Erwin Schrödinger, (one of my heroes) said in 1944 — before anyone knew what DNA was — that it must be an aperiodic-crystal. He gave a series of lectures, which later became the famous booklet, What is Life? It can be purchased for fifteen bucks on Amazon.com.
Schrödinger’s booklet changed the world — it’s one of the most prophetic works I’ve ever read. The tract changed my world view anyway; my view of life certainly.
It turns out, Schrödinger was right. DNA bundles store billions of bases in more-or-less random — but frozen — sequences much like crystals.
Just as molecules arrange themselves inside crystals, the bases inside DNA molecules also have an order, yes, an arrangement for sure, but it’s not a code; it’s not even a cipher; it’s merely a starting point for the most chaotic, complex, and messed-up process in nature — the creation of thinking, speaking, conscious-life (and less capable life) — all formed from a relatively few not-so-simple materials.
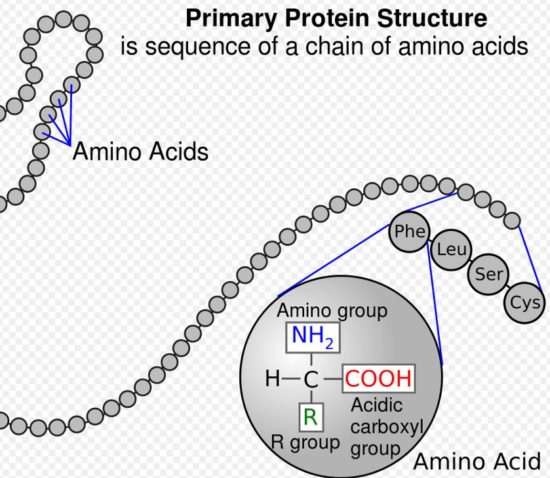
Here’s another assertion that might be difficult for some readers to accept. Genes don’t really exist. There are no free-standing genes; certainly not in human DNA, anyway. What scientists call genes must be constructed; they must be built; they must be put together; they must be fabricated, collected, and transported by molecules called RNA and by other processes known collectively as epigenetics. More on epigenetics later.

Most graphics and videos on the Internet seem to buy into a tidy notion that DNA is a code (not a reservoir and a starting point for the fabrication of templates). This notion seems to demand laser-precision and machine-like twelve-sigma reliability during protein synthesis.
Don’t believe it.
Yes, there is no argument; we can improve our chances for healthier lives by cleaning up less-than-optimal base-sequences — which are, as often as not, scattered, scrambled hodgepodges — using, hopefully, gene therapies like CRISPR. (Clustered Regularly Interspaced Short Palindromic Repeats)
But until technicians create tools to deal with other processes; until biologists can manipulate RNA itself and learn to change the ”weather” patterns (discussed later) inside cells, physicians will not be able to eliminate many of pathologies that plague our species and lead to diminished health and, for some, death.
RNA in all its forms (and there are many) is itself constructed from scattered templates that are hidden haphazardly like Easter eggs within the billions of bases strewn along the dozens of spirals inside a DNA bundle. RNA first builds itself up by interacting with various sections of base sequences in the DNA and then copies those sequences by borrowing matching freebases, which are floating everywhere in the medium of the cell’s nucleus.
RNA is much shorter in length and less stable than the DNA it models. But it doesn’t mutate as much as folks might expect, because it is also shorter-lived and reproduces less often. It’s more versatile too; more agile, because it is single-stranded; DNA is double-stranded.
RNA, in all its forms, is the workhorse of cell functions; it is both the building material and the construction machinery used in many important cell structures, which perform the yeoman’s task of protein building inside cells.
It seems plausible to me that over a few hundreds-of-millions of years the possibly self-generated RNA sequences may have acquired — through accident, luck, or trial-and-error — the ability to select, copy, paste, and assemble short sections of random DNA bases, which every-once-in-a-great-while actually worked to help build useful proteins that added survival advantages to their evolving hosts.
Maybe RNA designed and built DNA in the first place, which it learned to copy and manipulate. We may never know exactly how.
One thing scientists agree on: one-celled life was already highly developed, complex, and flourishing by the time the new planet, Earth, reached its first billionth year. Earth is four-and-a-half billion years old. Life came on fast during extreme conditions vastly different than now. This fact is amazing. No one understands how.
It has taken an additional three-and-a-half billion years to get to humans and the space-traveling civilizations that seem to dominate the earth today.
Thinking about RNA and DNA can be a frustrating circular process, much like the chicken or the egg problem; which came first? My sense is that most scientists today believe RNA came first, DNA later. Inside our cells, it is impossible to tell, but there is no denying that RNA’s diversity and flexibility make it a most likely candidate.
Many kinds of RNA live inside cells. Some run around doing nothing. They simply try to survive inside the complicated universe that is the typical living cell in every animal, plant, and microbe. They are called selfish RNA.
Most RNA sequences are much less selfish. They are like Christmas elves who work day and night; some to open Santa’s bags to gain access to their contents; others to copy various sections from the strands of blocks inside; others to move the copied sections to an assembly area, where other elves glue the copied segments together to form new sequences — many of which, by the way, are very different from the original sequences that the RNA elves found inside Santa’s gift bags; inside the DNA.
Eventually, messenger elves transport the long strands of little blocks they copied and assembled; they move them away from the center of the cell; out to the gooey regions of the cell beyond its center where other transfer elves are busy assembling (by threes) free-floating blocks (called bases, remember) and attaching these triplet-blocks (called anticodons) to single amino acids. The resulting structures are called transfer RNA (or tRNA, for short).
An amino acid is simply a configuration of carbon atoms with amine (ammonia) stuck to one side and carboxylic acid (vinegar) stuck to another — plus some other simple stuff attached here and there to make each amino acid unique among all the others. Think of an amino acid as a colored necklace bead. Out of the five-hundred or so differently colored beads in nature, transfer elves in humans work with only twenty or so.
Stay with me now. You just read the most difficult sentence in the essay. These amino acids attach themselves like colorized necklace beads to triplet-blocks (called anticodons) according to which of the three blocks (or bases) the transfer-RNA (tRNA) is made from. Watch the video ”From DNA to protein” above to better understand.
In the meantime, while all this other stuff is going on, the messenger elves are directing their long strands of copied-and-pasted blocks (bases) away from the cell’s tiny nucleus (center) toward little triplet-body-handling factories (called ribosomes; ribo for triplet, soma for body), which live in the inner goo (the cytoplasm) of the cell.
Many ribosomes are attached to a winding ribbon-like structure called the rough endoplasmic reticulum. Endo is Greek for inner; plasma is goo; reticulum means network.
At the same time, transfer elves in the goo (cytoplasm) steer their three-blocks-plus-a-colored-bead assemblies — in humans these “three-base” combos and twenty or so colored beads can be arranged forty-eight ways — into the ribosome factories, where they are matched-up to the blocks (bases) in the long strands that are being delivered from the cell’s nucleus (like cars in a choo-choo train) by the messenger elves.
Inside the ribosome factories, the triplet-blocks-plus-one-colored-bead assemblies, which have been constructed and collected by the transfer elves in the cytoplasm, are paired block for block (that is, base for base) to the long train of blocks that were collected, arranged, and carried by the messenger elves from the cell’s center (its nucleus).
As each transfer assembly triplet is matched-up three bases at a time to the blocks in the long messenger train, the single amino-acid bead that the three-block transfer assembly carries is ejected out of the ribosome factory.
Assembly elves — think of them as molecular forces — secure each ejected amino acid bead to the next bead, one after the other — in the exact order demanded by the order (in threes, called codons when located in the messenger RNA) of the bases (blocks) in the messenger sequence — creating as they go an amino-acid-chain, or necklace.
Once the amino acid chain (necklace) is long enough (and remember: there can be as many as twenty-three different colors of amino-acid beads in each necklace, and each necklace can be almost any length at all — up to hundreds or even tens-of-thousands of beads long) elves (molecular forces that work through the micro-scaffolding of the cell) go to work; they transfer the chains to Golgi structures, where they are bundled and folded into the twisted shapes that make them proteins. Like an Amazon distribution center, the Golgi apparatuses deliver the proteins to their destinations.
Like holiday elves, they deliver protein toys to every child’s bedroom in the cell, which in this analogy lie inside the abyss of the cell’s cytoplasm (cyto means cell; plasm means goo).
Some elves might feel compelled to deliver their proteins down the street to other homes (cells) in other neighborhoods by way of certain processes known as cell migrations.
These migrations can bring healing to injured tissues in other parts of the body, among other benefits. However, as I wrote earlier, stem cells that live inside the tissues of the body do the heavy lifting of cell replacement and healing.
Here is a way to visualize a human cell: think of cytoplasm (the cell goo) as the yolk of an idealized egg. A chicken egg is nothing like a human body-cell, but it makes a good model for explanations.

The nucleolus (at the epicenter) is a tiny, hard to find collection of proteins, RNA, and DNA at the very center of the nucleus where ribosomes are fabricated, if anyone wonders.
Ribosomes are made entirely from RNA; they are, in fact, one of the most ancient structures in cells; they are essential players in both prokaryotic and eukaryotic (ancient and modern) cells, which I will discuss in more detail soon.
Chromatin (the usually unorganized mess of DNA) lies in the nuclear goo of the nucleus that surrounds the nucleolus.
Remember that ribosomes are the tiny factories, where proteins get their start. Ribosomes themselves move between the nucleolus and the surrounding nuclear goo until they are ejected out of the pores of the nucleus into the goo of the yolk (which in this analogy is the cytoplasm) where many will float freely in an ocean filled with a dozen or more other structures important to cell functions. Many ribosomes attach themselves to the endoplasmic reticulum.
But we’re not concerned about these other structures right now. Proteins are the most essential substances from which our bodies are made.
Some biologists believe that as many as 50,000 different proteins are required to construct a human being. Others say 100,000. The human body is capable of producing two million. Each cell type on its own is capable of making 5,000.
Ribosomes are very important, because it is inside the ribosomes where proteins get started, so we concentrate on them first.
Think of the cell’s membrane as the “white” of the egg. It surrounds and protects the vital cytoplasm, where the making of proteins takes place.
Of course, the chicken egg analogy had to break down. A chicken egg is surrounded by an oxygen-permeable calcium carbonate (CaCO3) shell. Forget about the shells of chicken eggs. Human soma-cells aren’t protected in quite the same way.
In a chicken egg the nucleus is a little white mass that sits on top of the yolk and feeds on it. The nucleolus is inside that little white mass. So the breakfast-egg analogy falls apart pretty darn fast. It might be more confusing than helpful. I hope not.
A lot more is going on. And — I have to say this — the cells of most microbes (one-celled life, like bacteria and archaea) don’t look like the sunny-side-up cells of humans or most other animals and plants. For one thing, they are a lot smaller.
Bacteria and archaea can be from 20 to 10,000 times smaller than the eukaryotic cells of animals and plants. They lack membrane-bound organelles; they lack a nucleus. They look more like little sandwich bags of loosely cooked, scrambled eggs. Scientists call them, prokaryotes. (It’s Greek, meaning before they became fully formed kernels.) They are the ancient cells.
As for the other structures that live inside our own eukaryotic cells (again, it’s Greek, meaning after they became kernels) — they make a fascinating study, but are beyond the scope of this essay.
Eukaryotes are the modern more advanced cells of all plants and animals. It took two billion years for ancient prokaryotic cells to evolve to the modern eukaryotic cells that first appeared 1.5 billion years ago; it is these cells that congregated and evolved to become the plants and animals of today. Click on the links in this essay — such as the links in nearby paragraphs — to access Wikipedia articles, YouTube videos, and other sources to learn more about them.
Why is DNA called nucleic acid?
Phosphoric acid, which people have used for centuries to remove rust and to fertilize crops makes the DNA supports (or strands) on which hang the rungs of billions of bases. Phosphoric acid is the concentrated, clear syrup that makes Coca-Cola sting the tongue (carbon-dioxide bubbles make Coke sparkle). It’s the acid found inside the nucleus of every cell.

Hundreds of years ago phosphoric acid was made from the stone mineral apatite (calcium phosphate) and sulphuric acid — by-products of the mining and smelting of ores. Today the production process is more arcane and efficient.
Phosphorus is arguably one of the most important elements of life. Only magnesium is more important to viability, because the ATP that powers all cells will not work unless it is bound to it. Folks who aren’t eating whole wheat, spinach, black beans, almonds, and peanuts should probably be taking a daily magnesium supplement.
People who eat steak consume goodly amounts of phosphoric acid. Early researchers always found this acid in the center of cells, their nucleus. Scientists called it, nucleic acid.
Much later scientists discovered that the DNA sacks were full of bases, crammed together into tangled masses of long, curly chains they called chromosomes (Greek, for colored bodies).
Chromosomes stained well during lab experiments, which was fortunate for researchers, because color made it easier to see the chromosomes during the short periods of time when the genetic material in the cell’s center took on its distinctive form from out of the shapeless, invisible chromatin, where it lived.
It is by a curious twist of chemical engineering — to my mind, at least — that the bases don’t react with the phosphoric acid that anchors them. The DNA molecules don’t collapse into little piles of salt, like one might expect. Maybe they should; but on Earth, they don’t.
In this sense — the sense in which our DNA is made from acids and bases — we are salt, or could be; we are potentially a very complicated salt, yes, but a salt nonetheless.
A strong argument can be made that proteins and the polypeptide chains that make them are in fact salts. For reasons known only to biochemists, naming conventions hide this reality.
Amino acids, polypeptides, and proteins are thought of as inner salts and given the name zwitterions, of all things. Salt is at the heart of what and who we might become were it not for idiosyncrasies of nomenclature and the miracle that makes life live.
But to get back to phosphoric acid…
Phosphoric acid (phosphate) sacks (or strands) are loaded with toy blocks (bases), so they need something sticky, like sugar, to keep the blocks from falling out. The D in DNA stands for the sticky stuff — deoxyribose, which means sugar.

DNA is deoxyribonucleic acid.
The sugar and acid, together, form the rails of the famous spiral staircase, upon which the rungs of bases are hung. It’s the most incredible structure in nature. It’s called the double-helix. Some people named Watson, Wilkins, and Crick won a Nobel Prize for figuring it out.
To give readers a sense of scale: If someone were to take the longest strand of the double-helix in our DNA (it’s in chromosome-one) and somehow increased its diameter to one-inch (the thickness of a large garden hose), the DNA strand — when pulled straight — would increase in length to 567 miles (about 40 miles longer than the distance between Nashville and Detroit).
The bases (or toy blocks, as we’ve been calling them) would stack in pairs, eight-pairs-to-the-inch, along the entire 567-mile length of the hose. In this single chromosome, each of the nearly half-billion bases it contains (in 247,199,719 base-pairs) would be about the size of a Tic-Tac breath mint; maybe a bit smaller.
At normal scales, the dimensions are too fantastic to believe. A solitary strand of DNA can’t be seen, even with the aid of most microscopes, but if all the DNA in a single cell could be laid out end to end and flattened to remove the kinks, it would stretch to six-and-a-half feet. 6.5 feet is the length of all the DNA in a typical human cell.
According to Siri, 1E14 cells make a human. It’s “one” followed by 14 zeros. It’s 100 trillion. Other sources say no; the number of cells in a human being is between 15 and 70 trillion. A trillion has twelve zeroes, right?
Do the math. It will show that all the DNA in a single human is enough to spin a strand from the earth to the sun and back 99 to 662 times (which is somewhere between 198 and 1324 astronomical units, right?) depending on who is trusted to do the human cell-count. Does anyone believe it?
An astronomical unit (AU) is simply the distance from the Earth to the Sun. It is the distance traveled by light during 499 seconds, which is 8 minutes and 19 seconds. It’s close to 93 million miles, right?
The point is this: strands of DNA are too thin to see, but in humans their total length is on an astronomical scale that spans two-and-half to sixteen-and-a-half times the diameter of the solar system out to Pluto — a diametral orbital distance that is nearly 80 AU.
It’s a lot of DNA, even if the exact amount is uncertain.

Rosalind Franklin, the gifted X-ray crystallographer, who did the experimental research that led to the discovery of the double helix, died of ovarian cancer at age 37. The Nobel Prize Committee has a long-standing policy of not awarding prizes to people who have died. It’s why Irish physicist and mathematician John Stewart Bell didn’t receive a prize for his civilization-changing work on quantum-entanglement, after he suffered a brain hemorrhage and perished in 1990.
In Franklin’s case it was doubly sad, because she was also doing important work on the molecular structure of viruses related to polio (funded by the United States Public Health Service) when cancer overtook her. Once again another scientist — this time her partner, Aaron Klug — received the Nobel Prize that she might have shared.
Had Rosalind Franklin survived to receive two Nobel Prizes — one for her work on the double helix and the other on the structure of polio virus — she would be a household name, like Albert Einstein or Francis Crick or Jonas Salk.
Rosalind lived in a generation and a culture that devalued her; she was a woman who competed with men, some of whom may have undercut her and wanted nothing to do with her, a few admitted. It was a different time, the 1950s. Who knows where the truth lies?
For Franklin, fame-and-fortune wasn’t to be. Blame cancer, a disease of the “genes”, which she sacrificed her life to understand by working daily with the deadly X-rays that helped her unlock the secrets of viruses and, most important of all, to finally pull aside the opaque curtain that was hiding the shape of the molecule of life: DNA.
Once the structure of the double-helix became known, the potential to store information in a molecular bundle constructed like DNA was immediately recognized — and it appeared to be unlimited. It is why everyone at first thought that the DNA molecule must be a code, like an old-fashioned computer tape.
I’ve suggested that DNA is not a code; neither is it a cipher. Some researchers view DNA more as a storage device and a starting point for processes — complicated processes — that have taken place inside every living cell for 3.5 billion years.
Yes, a group of three bases and an attached amino acid, properly transformed and manipulated by RNA elves inside little protein-making workshops called ribosomes, can help to fabricate and string together colored beads to make necklaces (chains). Chains of amino acids (polypeptides) — properly ordered and folded, again by RNA elves — can become proteins.
Scattered DNA base sequences inside a cell’s nucleus, its center, are a starting point for an involved and complicated process of selection, duplication, transformation, and fabrication before anything useful can happen; before proteins can be built and released for living.
To think realistically about life, especially human life, people should remind themselves that two-thirds of their bodies is water; two-thirds of what’s left is protein; the rest is mostly fat.
Proteins are critical. Unless proteins are made right and duplicated accurately, life-forms will drift; life will change — as it certainly has over the 3.5 billion years that cellular life is known to have lived upon the earth.
The interior of a cell is a complicated space. The space between cells is other-worldly.
The process that goes on inside cells, instead of being thought of as a precisely executed computer code, might better be compared to the process of weather found on every planet in the solar system.
Each planet can be identified by its surface weather, which starts from a kit of basic materials, and is amplified by an avalanche of environmental conditions and chemistries, much like the bases in chromosomes, which are selected, copied, shaped and reshaped, configured and reconfigured by RNA elves and other characters we have yet to meet (because the science of the evolving genome — the genetic material — and the phenome — what animals and plants look like — is still young, and scientists understand less than the little they think they know about the complete process, at least so far).
The processes used to construct life forms by starting with the bases in DNA are analogous to the processes astronomers observe on the planets of our solar system, where each planet creates its weather from the matrix of materials and thermal conditions that seems to define it. Each planet in the solar system has a characteristic weather profile that depends on a chaotic interplay of materials and environment unique to that planet.
Earth has weather; so does Mars and Jupiter. Those who study planets know immediately which planet is which, simply by observing its weather. From telescopes on Earth, each planet looks like its weather. Each has its characteristic colors and patterns. Weather is a planet’s phenotype; it’s what folks see when they look.

That’s how it is with life forms, too. Each life form is the result of weather patterns inside cells, which give each animal, plant, and microbe its unique essence; its physical presence in the larger world where it lives.
It becomes conventional wisdom to think this way about life forms, when one considers that identical twins — two humans who share exactly the same DNA — always display a variety of differences when examined closely.
Identical twins never have the same fingerprints, for example. There are systems of weather occurring around their genetic material in every protein-producing region in their bodies. Epigenetics is the technical term for the study of how it is that variations in phenomes occur in organisms that have identical genomes — that is, identical gene sequences.
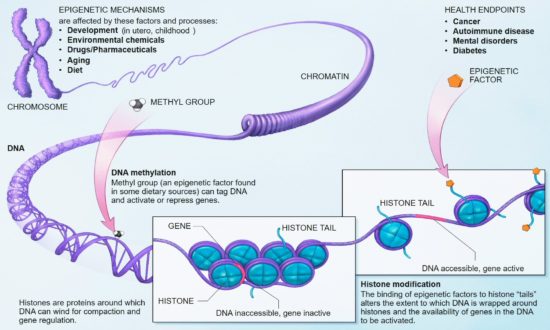
The outcomes of these storms are never the same; two supposedly identical children do not always receive the same toys from the RNA elves who rummage through their shared bags (strands) of DNA for the bases they will copy and rework into proteins. Things get mixed up and turned around. One toy gets selected; another doesn’t; one is painted green; another purple.
What I find interesting is this: identical twins get less identical as they age. They are easier to distinguish.
How many of the differences are due to variations in the production of proteins, which occur at the molecular level?
How much of the variation is induced by external stresses on the genome caused by lifestyle differences? How much is driven by an unavoidable drift in the statistics of protein production, which emerge and diverge over time in each phenome of the twins regardless of their lifestyle choices?

It’s a chaotic process that produces life on the earth. It’s a process that cannot be described or predicted by mathematics. If it could, scientists might take the DNA from prehistoric bones to create the original animals. Jurassic Park would be more than a Hollywood fantasy, which is what the book, movie, and sequels were and are.
An animal cannot be constructed from its DNA alone. A lot more is required than a simple collection of sequences formed from four bases and frozen in a molecule of DNA. A lot more of life’s machinery is required. The RNA elves — millions of them, like colonies of ants — must do their work.
When the work is done, and a protein has been made and delivered, the path back is lost, forever. No way exists — or is even possible — to reconstruct the sequence of bases in the DNA that started the process that built the protein. The process is not backward compatible, according to Matthew Cobb, the British zoologist and historian, in his latest book, Life’s Greatest Secret. Not only are the DNA sequences not reachable from knowledge of the proteins alone, but the processing steps that took place between the protein and the DNA are unknowable.
I don’t want to get too Mathy but think about this: sixty-four three-block (or three-base) sequences can in theory “code” for a mere twenty or so amino acids. It means that as many as three or four of those three-base sequences (the transfer elves we talked about earlier) can “code” for the same amino acid.
Reminder: four bases are all the choices DNA offers. Taken three at a time, they are more than enough to “code” for twenty or so amino acids. Get out the calculator, those who don’t believe it. 4 X 4 X 4 = 64. As mentioned earlier, humans have acquired over the eons 48 three-base combinations to work with. Bacteria, for another example, have 31 according to Matthew Cobb. We need less than two dozen.
Amino acid sequences long enough to form proteins can be hundreds to tens-of-thousands of acids long.
Forget about how amino acid chains get folded properly to make proteins. How can anyone work backwards from a protein formed from thousands of amino-acid beads — each one of which was secured to any one of three or four different 3-way combinations of bases (or blocks) — and then go about the task of reconstructing from all those possible combinations the exact sequence of bases (or blocks) in the original DNA, which more than a few random RNA sequences interacted with to make their choices from billions of bases in the first place?
Take a breath. There isn’t enough time in the history of the universe to figure it out for the tens-of-thousands of proteins that it takes to make a functioning animal or plant.
Raise the number “three” (or four, or five, or six, or two; it doesn’t matter) to the thousandth power on a calculator, those who may be having trouble accepting a possibly demoralizing fact. Most calculators will spit out the word, OVERFLOW. The number of possible sequences is impossibly large. It might as well be infinite.

It seems likely that the ”code” has changed in dramatic ways since the first primitive cells formed 3.5 billion years ago. British zoologist, Matthew Cobb, has suggested that words like ”code” and ”information” might better be thought of as mere metaphors when applied to the machinations of complex molecules like DNA and RNA, which — can we admit? — operate on quantum scales for which we humans have no natural intuition.
There are thousands of chains of all different lengths and folding patterns. No one is going to reverse-engineer the DNA of a life-form as complex as a human being from its proteins; nor from its RNA elves; nor from its essential enzymes and catalysts; not anytime soon; not ever. It goes for dinosaurs, trees, or any other reasonably complex living thing — now or from the distant past.
Why do we have to reverse-engineer? Why not read the instructions right off the DNA itself? By now most readers must be starting to understand that the sequences necessary to build proteins are scattered among billions of bases. We can’t find the right ones in the right order. It’s not possible; not for creatures as complex as humans or dinosaurs.
Even if someone could reverse-engineer DNA sequences from proteins, how would they construct and organize the ant-like colonies of RNA elves that must sort through the DNA bases to select and build the right sequences; how do they identify and isolate the sequences necessary to build and orchestrate, for example, the tens-of-thousands of enzymes that are required to give researchers any chance at all to build a functioning human-being or even a prehistoric dinosaur?
It gets more complicated.
Those who don’t believe it might want to read about CNVs, or copy number variations, that disrupt the probabilities that certain base-sequences in genes can be fabricated in the same way time after time. Click the link.
It’s a form of change that has nothing to do with mutations or inheritance. It is more related to the untidy mess that three billion base pairs make whenever they get together to do anything at all. It’s a genetic Woodstock of variation, where almost anything can happen and sometimes does.
Now might be a good time to mention that there are virus infections that can alter the DNA in cells. These viruses are called retroviruses, because they reverse the DNA to RNA transcription process described earlier by introducing an enzyme into the cells they infect. This process is disease producing — it causes cancer — and destroys the host animal (or person) if left untreated.
The enzyme, reverse transcriptase, has become a tool that molecular engineers now use to modify organisms in experiments. Enough said. The weeds of molecular biology grow thick and deep.

Tables like the one above make the ”code” appear comprehensive and far-reaching. Some readers might be surprised to learn that only one or two percent of the bases in human DNA are ever copied by RNA to make proteins.
Added to 2% for protein synthesis is 8% to make little pieces of RNA that oversee and coordinate the process of protein-making — like colonies of swarming ants. The rest of the bases (90%) do other things; maybe — many of them — do nothing at all. No one is really sure what they do or don’t do.
Geneticists used to believe they could clone animals from their DNA sequences alone. Yes, there once was a cloned sheep named Dolly. Some readers may have read about her.
After many heart-breaking failures, researchers managed to take the DNA from the mammary gland of one sheep, inject it into the nucleus of an egg from a second sheep, and implant this DNA cocktail into the womb of a third. By some miracle, Dolly was conceived and born, on July 5, 1996.

Sheep live twelve years, on average. By Dolly’s fourth year arthritis crippled her. At year seven researchers euthanized her; she had developed a chronic and incurable lung disease.
Dolly was the recipient of arguably the best healthcare any sheep ever received in the history of veterinary medicine. She didn’t do well. Click this link for an update on the Dolly research.
For the sake of complete accuracy, permit me to admit that no one I know clones sheep anymore. There are too many failures. The failure rate for clones is right around 100%.
Some breeders claim to clone horses, which they sell to folks who hope to increase their odds of winning races. I can argue that their definition of cloning differs from science, and the proof is that the horses have noticeable differences which negatively impact their ability to win.
Clone researchers, as far as anyone knows, have never used DNA alone, anyway. All borrow the enzymes, RNA, ribosomes, and other cell structures of other life forms to incubate the DNA they play with to try to create “artificial” life.
Mitochondrial DNA is unreachable. It is produced only in the ovum of the female. Mitochondrial DNA carried by sperm is miniscule in amount and quickly identified and destroyed in the fertilized egg.
Any technique that involves in vitro fertilization can bypass this natural process and inadvertently scramble the DNA in mitochondria. It can be debilitating, even disease producing.
Any technique that swaps out nuclear DNA while avoiding mitochondrial DNA doesn’t get to the power source of cells — a major enabler of stamina and endurance. In racehorses, the role of mtDNA is probably crucial, it seems to me.
My point is that duplicating a DNA sequence is not enough to produce an identical copy of an animal as complex as a sheep, horse, or human. Too much other stuff is going on during reproduction that is not controllable or even known.
And speaking of enzymes, can we please not go there? I’m reminded of Chris Farley in the 1992 movie, Almost Heroes.
Does anyone remember?
His tutor asks him to learn the symbol for lowercase A. ”What do you want from me?” Chris bellows while rolling his eyes and clawing his hair. ”You want my head to explode?!”
Well, no, of course not. But for those who have to know more, why not push ourselves just a little bit harder? May I point out the obvious? Enzymes speed up chemical reactions. A chemical reaction that might under normal circumstances take years can be reduced to milliseconds by an optimally configured enzyme.
Some enzymes are made from RNA; most are proteins; in fact, most proteins are enzymes; they all get their start from sequences of bases hidden deep within the mountains of DNA inside our cells. These bases are selected, copied, and transformed into their many convoluted shapes for a very special reason: to help accelerate over 5,000 processes inside cells.
Without these highly specialized structures, metabolism would grind to a halt; DNA and RNA would acquire all the mobility of a conga-line of standing stones; cell processes would freeze into a petrified forest of non-living complexity. Life as we know it would be impossible, code or no code.

Here is a good question: Has any research team ever created artificial life in a laboratory?
Craig Venter, who has been interviewed on 60 Minutes and appeared in several Ted Talks videos, says that he has. He oversees a number of research labs funded by big oil and the government. His labs write computer code to generate base sequences, which they construct and then inject into yeast (among other techniques) to produce life-forms that they hope will someday lead to biofuels and greenhouse gas inhibitors.
Among other accomplishments, one of the labs, the Craig Venter Institute, is known to have introduced a gene from the bacteria, escherichia coli, into the earth’s toughest microbe, “Conan the Bacterium” (Deinococcus radiodurans), to create microbes that can detoxify radioactive wastes at nuclear facilities.
So, the answer to the question about whether anyone has ever created artificial life must be, probably not, not really — not from scratch, anyway. Yes, people have done amazing things. No one has created life without using existing life to do it, though. The process is too complex. On Earth, it has taken 4.5 billion years.
Many argue that life fell to Earth from the stars. Even Earth itself might not have been able to ignite the spark that led to humans and all the life we know.
In 2012 a different group of researchers did find a way to arrange a set of different bases inside DNA-like molecules called XNA. But it was a way of coding sequences only; it didn’t produce or even arrange proteins into anything that could be called, alive.
The “X” stands for the Greek word, Xeno, which means “other.” XNA is other nucleic acid.
An informed reader told me that in fact a protein was made from a sequence of XNA in 2015. If true, the future of genetics could get interesting in coming decades.
XNA is at the very least the precursor, many hope, for long-term storage of massive amounts of information in small, stable molecules — demanded now by data-churning behemoths such as CERN, home of the world’s most powerful particle-collider, located in Geneva, Switzerland.
Just because artificially constructed molecules like XNA can store useful information does not mean that DNA does the same. People have imagined meaning into the bases of DNA, which they simply don’t have — to help better understand their function and to more effectively manipulate them — for good or ill.
Another development in 2012, which some readers may remember, is that researchers learned to use a process known by the acronym CRISPR to change the sequence of bases in stretches of DNA. They adapted an immunization process that bacteria use to kill viruses and defend against subsequent attacks.

Bacteria use CRISPR to suck the DNA out of an attacking virus, which they store in a kind of library for future reference. If a bacterium survives and the virus dies, somehow the bacteria is able to develop a quick-kill strategy that it will use whenever it is invaded by more DNA that matches a copy in its collection.
Researchers learned to create novel CRISPR DNA based on the system used by bacteria. They then attached RNA guides and cas9 protein shears to the sequences. They learned to deploy the assemblies to search and destroy bad DNA; and to insert designer DNA in its place in the cells of plants and animals; even humans.
These scientists insist that gene “therapies” are necessary, because the fact is: DNA is defective — most of it, anyway. Very few humans are symmetrical, attractive, disease-free, smart, emotionally stable, long-lived, or any other desirable trait anyone might want that is driven by how humans are built, or how they are “coded” at a molecular level.
Crisper video published October 25, 2019 on YouTube.
Gene drivers (mentioned in the first paragraphs of this article) are being developed in coordination with CRISPR techniques to enable changes to DNA molecules that will be permanent and transmittable 100% of the time. Their success will depend on how well lab technicians understand what is going on inside the molecules of life; and inside our cells.
Editor’s note: In January 2018 some researchers admitted that problems related to positional locating have created a roadblock to success for CRISPR technologies. They hope to solve the difficulties soon to avoid a catastrophic failure in the application of this heralded gene-altering process. One process under development that seems to promise more precision and speed is to use electro-magnetic positioning in place of viruses.
I believe we need to slow down and learn more before we unleash immortal genes into the biosphere that no one can pull back and which may turn out to be harmful despite best intentions. Asilomar style conferences that lead to best-practice regulations with the force of international law behind them are desperately needed to control biotechnologies that are quickly getting out-of-hand and beyond the control or understanding of government and politicians.
It is quite certain that PCR technology (polymerase chain reaction amplification), which scientists use to amplify into a viewable goo the molecules of DNA-style life might be misleading folks into believing that DNA-style life is all there is.
Earth could be infested with non-DNA based life, but no one will know until other technologies capable of detecting and amplifying it are developed and perfected.
People need to remind themselves that we are talking about molecules here — molecules of life that can’t be seen — even with help of the most sophisticated microscopes. Everything science knows comes from amplification techniques and mathematical analyses. I hope someday to write an essay on the techniques scientists use to tease out what they know for sure about these next-to-impossible-to-observe molecules.
Serious scientists refer to the possibility for the existence of non-DNA style life as the “shadow biosphere.” If this non-DNA life interacts with our own in a symbiotic way, the potential for harm, it seems to me, increases the more lab technicians play around with molecules they don’t fully understand while they remain oblivious to life they can’t detect, because they lack appropriate laboratory tools and techniques.
An even messier problem is “dark DNA”. It’s DNA that can’t be found, though tests clearly show it must exist for certain cell processes to work right.
Some researchers argue that as many as twenty thousand proteins are manufactured in humans that, when they search the human genome, they can’t find the sequences that are required to be there, somewhere, to enable the proteins to be built. I urge readers to click this link to learn more about this potentially serious inability of sequencers to decode DNA accurately and completely.
No one knows what they don’t know; and what they don’t know can kill us all, if lab workers aren’t cautious. Researchers know they don’t know stuff — important stuff if they intend to play around with gene drivers and CRISPR induced gene sequencing.
Researchers might be walking through a genetic minefield but are so eager to cross that they ignore the dangers of amputated limbs; the loss of sight and hearing; the possibilities for disfigurement to the genomes and phenomes of species like our own, which all people may one day come to regret.
No human is perfect. Sometimes our imperfections are caused not by bad stretches of DNA but by naughty RNA elves who copy less than optimal sections of bases, which they hammer together into less-than-optimal genes, which can screw-up a sequence of amino acids. The RNA elves end up making defective proteins that pollute cells, damage our bodies, and make our lives miserable.
To the extent that these screw-ups are the result of a lousy sequence of bases in our DNA, perhaps these patterns will be able to be altered using CRISPR technology (if anyone can get it to work right), which is likely to increase the odds of inducing better outcomes. But many screw-ups, perhaps most, are not caused by poorly sequenced genes constructed from DNA.
Many problems result from bad choices made by some arbitrary RNA elf, for example, who might have decided, perhaps, to cut and paste a random mix of bad sections it rummaged from the DNA strands; its errors and mistakes might not always be able to be located, identified, and repaired successfully. Renegade RNA elves are hard to track down and kill; at least so far.
Some problems can be caused by all kinds of things not related to DNA, like temperature, quantum effects, and cosmic radiation, including sunlight. The number of things that can go wrong with the weather-environment inside cells is enormous. Copy number variations in gene sequencing is another problem area that I mentioned earlier.
Safety and reliability are probably the two most important reasons why our haystacks of six-billion DNA bases hide a mere twenty-one thousand so-called genes, most of which are scattered in pieces throughout our vast DNA bundled-network.
Those few sequences that are important for survival are less likely to be attacked and mutated if they are surrounded by sequences of little or no value to survival and good health. Base-sequences essential to life hide within chromatin like proverbial needles in a haystack.

Big chunks of DNA are thought to be junk — relics left behind by billions of years of evolution and change. Junk DNA could be a legacy of screw-ups and obsolescence. Dolphins, for example, have noses, but can’t smell. They seem to have a lot of corrupt DNA sequences related to smell, which are broken and don’t work due to neglect and disuse.
Humans are no different. We have DNA we no longer use. Through disuse, our base sequences, some of them, get corrupted over time, some think, and become unusable. The base sequences don’t get up and go anywhere, though. They just hang around, paralyzed, doing nothing. They become unrecognizable to the RNA elves, who learn somehow to avoid them.
Mitochondria and bacteria don’t seem to have much, if any, junk DNA, but humans, like other animals and plants, have almost no nuclear DNA that isn’t junk. It’s kind of mysterious.
A Russian agronomist from the Soviet era, renown in his time as an expert on the cultivation of wheat, Trofim Denisovich Lysenko, believed that plants and animals which were unlucky enough to find themselves subject to environmental stresses could draw on reserves from a pool of what is today called junk DNA to change their hereditary direction and enhance their survival odds. His idea has yet to be discredited.
The simple onion has 16 billion base pairs in its DNA. The loblolly pine tree — it’s an important source of lumber, which thrives in southern swamps — houses 22 billion. Humans have 3 billion.
EDITORS NOTE: As of January 2018, a Mexican salamander that can regrow limbs (the axolotl) has been sequenced. It is known to have 32 billion base pairs.
What do all these bases code for? They code for nothing, apparently. Maybe they are a warehouse of survival tools left behind as the distant past of billions-of-years ago gradually transforms itself into now; our miraculous present.
Another compelling idea that occurred to me as I wrote this essay is that the tangled mess of unused DNA in every plant and animal might have grown both in volume and complexity during ancient times — quite apart from environmental pressures on the life-forms themselves.
Could massive DNA growth have preceded evolution to enable and accelerate biodiversity during unforeseen environmental catastrophes?
It’s important to find out, because statistical studies on the rate of mutations seem to support the idea that mutational frequency cannot be the primary driver of species differentiation. Mutation rates are too low; the process is a snail’s pace compared to what is needed to transform a chimpanzee for example into an orangutan; or primates of any kind into humans.
Is it possible that mammoth reservoirs of disorganized and unused bases grew and multiplied inside the nuclei of ancient cells — like molds in petri dishes — to fuel bio-explosions of diversity and complexity when conditions were right? It’s a thought.
An abundant supply of unused DNA combined with aggressive colonies of swarming RNA segments might help to explain rapid, diverse bio-blooms (and even account for absences in fossil records) that seem to have occurred during the Cambrian era — to cite one example out of many.
The world’s smartest people are just getting started in the field of molecular genetics. Despite all that others have learned, much remains to know; more, much more, remains to discover and understand. Secrets hide in the complexity that are certain to better explain how biodiversity bloomed on planet Earth.
DNA bases are not a code, it seems to me; they are simply a platform for departing mRNA trains that, when properly coupled, can become assembly templates for chains of amino acids — complex assemblies of molecules that depend on very many processes and structures to have even the remotest chance of being transfigured by ribosomes into a seeming infinity of unlikely proteins — matrices of proteins and other structures, which have risen from the dust and the seas like the miracles of angels; an endless froth of bubbles; a deluge of structures that have over eons shaped the messy, sometimes ugly, often beautiful human beings and all other life on our planet; our home; our beloved Earth.
Billy Lee

RECOMMENDED BOOK:
In 2015, the University of Manchester zoology professor, Matthew Cobb, published an incredible book: Life’s Greatest Secret. Science celebrity, Brian Cox — in typically British understatement — labeled it, “a bloody brilliant book.”
Adam Rutherford, the British geneticist said, ”This is the definitive history of arguably the greatest of all scientific revolutions.”
Life’s Greatest Secret is a must read for anyone who is interested in the science and history of the human genome. We strongly advise our readers to buy and read this important book. Billy Lee has read it twice, marking it up each time with magic-marker and margin-notes. It is a science blockbuster; a fantastic book written in an engaging, easy-to-understand style.
Billy Lee wants to acknowledge and thank Professor Mathew Cobb for writing Life’s Greatest Secret, which helped to inspire this essay: NO CODE.
The Editorial Board